Abstract
In this study, we have developed an efficient method based on single-stranded DNA (ssDNA) aptamers along with silica fluorescence nanoparticles for bacteria Salmonella typhimurium detection. Carboxyl-modified Tris(2,2′-bipyridyl)dichlororuthenium(II) hexahydrate (RuBPY)-doped silica nanoparticles (COOH-FSiNPs) were prepared using reverse microemulsion method, and the streptavidin was conjugated to the surface of the prepared COOH-FSiNPs. The bacteria S. typhimurium was incubated with a specific ssDNA biotin-labeled aptamer, and then the aptamer-bacteria conjugates were treated with the synthetic streptavidin-conjugated silica fluorescence nanoprobes (SA-FSiNPs). The results under fluorescence microscopy show that SA-FSiNPs can be applied effectively for the labeling of bacteria S. typhimurium with great photostable property. To further verify the specificity of SA-FSiNPs out of multiple bacterial conditions, variant concentrations of bacteria mixtures composed of bacteria S. typhimurium, Escherichia coli, and Bacillus subtilis were treated with SA-FSiNPs.
In addition, the feasibility of SA-FSiNPs for bacteria S. typhimurium detection in chicken samples was assessed. All the results display that the established aptamer-based nanoprobes exhibit the superiority for bacteria S. typhimurium detection, which is referentially significant for wider application prospects in pathogen detection.
Similar content being viewed by others
Background
Bacteria Salmonella typhimurium (bacteria S. typhimurium) is a kind of food-borne zoonotic pathogen, which may cause food poisoning, acute gastroenteritis, and exogenous febrile disease and even lead to death [1, 2]. Especially, food poisoning caused by bacteria S. typhimurium is at the top of the bacterial food poisoning event list in the world [3]. Generally, bacteria S. typhimurium is mainly detected using morphological observation, immunological method, and molecular techniques nowadays [4–6]. Briefly, the morphological observation method only provides the shape, size, and dyeing property (gram positive or gram negative) of bacteria, which cannot provide the specific types of information of the bacteria the researchers look for. Although the immunological method, like enzyme-linked immunosorbent assay (ELISA), is a widely used method for bacteria detection, the procedure is complex and time-consuming. The PCR-based methods are popular at present, but they are easily contaminated and time-consuming sometimes. Therefore, to develop a fast and effective method for bacteria S. typhimurium detection to remedy the extant weaknesses is essential in a pathogen-monitoring program as well as clinic diagnosis.
In recent years, the aptamer has attracted tremendous interest because of its excellent features for pathogen detection. Aptamers are kinds of synthetic peptide or oligonucleotide molecules which can bind to the target molecule firmly. With advantage of high affinity and specificity, the aptamers are prominent for detecting all kinds of targets, such as tumor cells, bacteria, virus, proteins, some small molecules (ATP), and even metal ions [7–11]. Compared with antibodies, aptamers would be promising recognition elements for bioanalysis because of their high affinity and specificity with various kinds of targets, easy preservation, facile modification, and good stability.
For the application in bacteria S. typhimurium detection, several aptamer probes had been selected and characterized using whole-bacterium SELEX [12–14]. According to the structural analysis of a selected aptamer and measurement of affinity constant between the aptamer and bacteria S. typhimurium, the bacteria S. typhimurium aptamer is a 40-base single-stranded DNA with Kd value (6.33 ± 0.58 nM), which indicates the high affinity and specificity of aptamers [14]. In addition, the single-stranded oligodeoxynucleotide anti-S. typhimurium aptamer has a lower molecular weight compared to antibodies.
The fluorescent dyes are materials commonly used as an indicator in bioanalysis, while the deficiency of photobleaching severely hinders the range of their applications. With the development of nanotechnology in recent years, fluorescent nanomaterials play an increasingly important role in bioanalysis [15]. Many researches had revealed that the nanometer material shell surrounding the dye molecule could prevent the dye from photobleaching. In addition, a larger amount of dye molecules entrapped inside the silica matrix would play the role of a signal amplifier. Then, the fluorescent core-shell nanoparticles, like the dye-doped silica nanoparticles, have attracted great attention due to their other good characters with respect to photostability, surface-to-volume ratio, modification, cost, biocompatibility, and hydrophilicity [16–19]. For instance, targeted fluorescence nanoprobes are prepared by conjugating the desirable biomolecules (antibody or protein) to the surface of the dye-doped silica nanoparticles, which have been widely used for tumor cell recognition and separation, bacteria labeling, and DNA ultrasensitive assay [20–22].
Tris(2,2′-bipyridyl)dichlororuthenium(II) hexahydrate (RuBPY) is a kind of hydrophilic positively charged fluorescent dye, which shows the fluorescence emission wavelength (610 nm) with the excitation wavelength at 450 nm. As mentioned above, compared with the free RuBPY dye, the RuBPY-doped silica nanoparticles take advantage of resistance to photobleaching due to dye molecules with a positive charge entrapped stably inside the negatively charged silica matrix, which prevents the potential quenching substance approaching the dye molecules [23, 24].
In view of the extraordinary properties of aptamer and fluorescent nanomaterials for target labeling and signaling [25–28], we prepare novel bioprobes based on single-stranded DNA (ssDNA) aptamers and RuBPY-doped silica nanoparticles for bacteria S. typhimurium detection, of which the availability was accessed in a variety of conditions.
Methods
Materials
Triton X-100, tetraethyl orthosilicate (TEOS), (3-aminopropyl)triethoxysilane (APTES), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide sodium salt (Sulfo-NHS), RuBpy, and streptavidin were purchased from Sigma Chemical Co. (St. Louis, MO). Ammonium hydroxide (25–28 wt%), n-hexanol, cyclohexane, acetone, alcohol, N,N-dimethylformamide (DMF), and succinic anhydride were purchased from China Pharmaceutical Group Shanghai Chemical Reagent Co., Ltd. Bacteria S. typhimurium, Escherichia coli (E. coli) DH5ɑ, and Bacillus subtilis (B. subtilis) were obtained from China Center for Type Culture Collection. The rhodamine B isothiocyanate (RBITC)-labeled bacteria S. typhimurium aptamer was purchased from Beijing Biosynthesis Biotechnology Co., Ltd. The biotin-labeled bacteria S. typhimurium aptamer: 5′-biotin-(CH2)6-AGTAATGCCCGGTAGTTATTCAAAGATGAGTAGGAAAAGA-3′ and the biotin-labeled random sequence: 5′-biotin-(CH2)6-TGTCATGACCCGTAGGTAGTCTTAGAAGACTAGGCACGTT-3′ were synthesized at Shanghai Sangon Biological Engineering Technology & Services Co. (China).
Main Instrumentation
The size and uniformity of synthesized silica NPs were measured by means of transmission electron microscopy (JEOL, JEM100CXII, Japan). Photoluminescence was measured using a fluorescence spectrophotometer (F96PRO, China). Fluorescence image results were observed under an inverted fluorescence microscope (Nikon ECLIPSE TE2000-U, Japan).
Preparation and Characterization of Streptavidin-Conjugated RuBPY-Doped Silica Fluorescence Nanoprobes
Carboxyl-modified RuBPY-doped silica nanoparticles (COOH-FSiNPs) were synthesized by the method described by Arriagada et al., and the detailed procedure referenced to Cai et al. [29, 30]. In short, the RuBPY-doped silica NPs were prepared through the polymerization reaction of TEOS and NH4OH with RuBpy in the water-in-oil microemulsion, which was made of Triton X-100, cyclohexane, and water. Then, the RuBPY-doped silica was amine modified using TEOS and APTES. Afterwards, the generated floccule was deposited using EDC and Sulfo-NHS and washed with acetone after centrifugation. By washing and resuspending with DMF solution, the products were reacted with succinic anhydride under nitrogen gas for 24 h with continuous stirring. Then, the prepared COOH-FSiNPs were washed with water and resuspended in PBS for the next step.
Two milligrams of COOH-FSiNPs, 1 mg of EDC, and 2.5 mg of Sulfo-NHS were added to 1 mL of 0.1 M PBS buffer (pH 7.4). The mixture was then incubated for 15 min at room temperature with gentle shaking. After the reaction completed, 50 μL of streptavidin diluted in PBS at a concentration of 1 mg/mL was immediately added to the solution, following a 3-h incubation with gentle shaking at room temperature. The particles were washed with 0.1 M PBS (pH 7.4) and then resuspended in 1 mL of 0.05 % BSA for 1 h to block the free carboxylates. After the reaction completed, the prepared streptavidin-conjugated silica fluorescence nanoprobes (SA-FSiNPs) were washed with 0.1 M PBS buffer (pH 7.4) three times and then resuspended in 0.1 M PBS buffer (pH 7.4) for further use.
Application of the Nanoprobes for Bacteria S. typhimurium Detection
One hundred microliters of 10 μM biotin-labeled anti-S. typhimurium aptamer and 1 mL of bacteria S. typhimurium diluted in PBS at a concentration of 80 cfu/mL were incubated for 60 min at 37 °C with gentle shaking. After the reaction completed, the suspension of bacteria S. typhimurium aptamer complex was centrifuged and washed with PBS three times and then resuspended in 1 mL of 0.1 M PBS (pH 7.4). One hundred microliters of SA-FSiNPs was immediately added to the suspension. After incubation for 60 min at 37 °C with gentle shaking, the mixtures were centrifuged (6000 rpm × 5 min) and washed with 0.1 M PBS (pH 7.4) three times to remove the free SA-FSiNPs. The probe-bacteria conjugates were finally resuspended in PBS and then smeared on a slide glass for fluorescence microscope observation. The corresponding control group was treated through the same procedure with exception of the anti-bacteria S. typhimurium aptamer which was replaced by a biotin-labeled random sequence aptamer.
Streptavidin-Conjugated Silica Fluorescence Nanoprobe for Bacteria S. typhimurium Detection Under Multiple Bacterial Conditions
Three bacteria mixture groups were set in terms of the concentration gradient:
-
Group A: 80 cfu/mL bacteria S. typhimurium + 40 cfu/mL E. coli DH5ɑ + 20 cfu/mL B. subtilis
-
Group B: 40 cfu/mL bacteria S. typhimurium + 20 cfu/mL E. coli DH5ɑ + 80 cfu/mL B. subtilis
-
Group C: 20 cfu/mL bacteria S. typhimurium + 80 cfu/mL E. coli DH5ɑ + 40 cfu/mL B. subtilis
The three groups were incubated with 300 μL of 10 μM biotin-labeled anti-S. typhimurium aptamer for 60 min at 37 °C with gentle shaking. After the reaction completed, three kinds of bacteria mixture were centrifuged and washed with PBS three times and then resuspended in 1 mL of 0.1 M PBS (pH 7.4). Two hundred microliters of SA-FSiNPs was immediately added to the bacteria mixture. Subsequent steps were as described in the “Application of the Nanoprobes for Bacteria S. typhimurium Detection” section.
Fluorescence Nanoprobe for the Bacteria Detection in Chicken Samples
Five grams of minced chicken samples purchased from a supermarket and 100 μL of 1 × 103 cfu/mL bacteria S. typhimurium were mixed in the triangular flask containing 50 mL of bacteria culture media (Mueller Hinton Broth). The mixture was then incubated for 12 h at 37 °C with gentle shaking. After the reaction completed, the culture media were centrifuged (1000 rpm × 5 min) and washed with PBS three times. The chicken samples were resuspended in 10 mL of 0.1 M PBS (pH 7.4). One hundred microliters of 10 μM biotin-labeled bacteria S. typhimurium aptamer was then added to the chicken samples dispersed in PBS. After incubation for 60 min at 37 °C with gentle shaking, the chicken sample suspension was centrifuged (5000 rpm × 5 min) with PBS three times. The chicken samples were then resuspended in 5 mL of 0.1 M PBS (pH 7.4). Two hundred microliters of SA-FSiNPs was subsequently added and then incubated for 2 h at 37 °C with gentle shaking. After centrifugation (1000 rpm × 5 min) and washing, the supernatant was transferred into a centrifuge tube and centrifuged (5000 rpm × 5 min). The probe-bacteria conjugates were finally resuspended in PBS and then smeared on a slide glass for fluorescence microscope observation. The procedures corresponding to the experimental groups were performed in the control groups except that the ground chicken samples were incubated with bacteria culture media in absence of bacteria S. typhimurium.
Quantitative Analysis of Bacterial Cell Staining
Quantification of bacterial cell staining was performed using ImageJ software according to its standard manual.
Results and Discussion
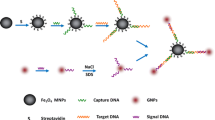
In this research, we used the reported biotin-labeled ssDNA aptamer specific for bacteria S. typhimurium as the recognition elements, combining the silica fluorescence NP as the detection probe. The anti-S. typhimurium aptamer is a 40-base ssDNA, which shows high selectivity and affinity to the whole bacteria S. typhimurium [14]. Owing to the potential strong steric hindrance effects between the bacteria and the rigid NPs, the implementation for the probe would be impeded. Then in our study, the biotin-labeled aptamer was not directly conjugated to the SA-FSiNPs to avoid the conjugate (aptamer-biotin-SA-FSiNPs) formation. Instead, the bacteria S. typhimurium were firstly incubated with the free biotin-labeled aptamer that could result in mass molecules bounding to the bacteria surface, which would facilitate the subsequent labeling of SA-FSiNPs. So the bacteria S. typhimurium suspension was firstly incubated with the biotin-labeled aptamer. As expected, the biotin-labeled aptamers would bind to membrane proteins on the surface of the cell walls of the bacteria. After centrifugation and washing, the bacteria were then incubated with SA-FSiNPs. The fluorescence nanoprobes could bind to the surface of bacteria S. typhimurium because of the interaction between the biotin and the streptavidin. As for the control groups, the biotin-labeled random ssDNA could not specially bind to the membrane target bacteria, which results in no fluorescence signal under the microscope. Therefore, the bacteria S. typhimurium detection result can be recognized on the basis of striking fluorescence feature on the surface of detected bacteria. The overall schematic is described in Fig. 1.
Characterization of Carboxyl-Modified RuBPY-Doped Silica Nanoparticles
The morphology information of synthesized silica NPs were measured by transmission electron microscopy. As shown in Fig. 2a, the NPs were uniform in shape with an average diameter of ~60 nm (the total number of NPs measured by the SIS image-processing software is 28, and the size of the measured NPs is 60 ± 5 nm). In addition, the fluorescence spectrum of COOH-FSiNPs was characterized by a fluorometer. As shown in Fig. 2b, compared with the fluorescence spectrum of free RuBPY dyes, COOH-FSiNPs have the same fluorescence emission wavelength (610 nm) with the excitation wavelength at 450 nm. Due to the mesoporous structure of silica, the RuBPY dyes entrapped inside the silica matrix are still excited [17, 31].
Labeling and Imaging for Aptamer-Based Bacteria S. typhimurium Detection
To confirm whether the prepared SA-FSiNPs were practicable as a bacteria-labeling probe, bacteria S. typhimurium solution was firstly incubated with biotin-labeled anti-S. typhimurium aptamer. After centrifugation and washing, the bacteria were then treated with SA-FSiNPs. Unbound nanoprobes were washed away with PBS. The probe-bacteria conjugates were then imaged using fluorescence microscopy. The procedures corresponding to the experimental groups were performed in the control groups except that the biotin-labeled random sequence was used instead of the biotin-labeled bacteria S. typhimurium aptamer. As shown in Fig. 3, a1 and a2 represent the bright-field and fluorescence microscopy pictures in the experimental group, respectively. b1 and b2 represent the bright-field and fluorescence microscopy pictures in the control group, respectively. The fluorescence images were taken with the same contrast and brightness. Compared with the bright-field picture in the experimental group, bacterial colonies with significant red fluorescence were clearly observed in the fluorescence microscopy image, which indicates that a large number of SA-FSiNPs were bound to the surface of bacteria S. typhimurium. In contrast, the control group shows no red fluorescence of SA-FSiNPs under the fluorescence microscope, which indicates that there is no nonspecific binding between the bacteria S. typhimurium and nanoprobes. Then, we could agree that aptamers could recognize and bind to targeted bacteria with high affinity and specificity. In addition, fluorescent labeling of bacteria is attributed to the interaction between the streptavidin on the surface of nanomaterials and biotin on the surface of bacteria rather than the nonspecific binding between the nanomaterials and bacteria [32–34].
Fluorescence images of bacteria S. typhimurium labeled with the biotin-labeled aptamer and SA-FSiNPs (A) and biotin-labeled random single-stranded DNA and SA-FSiNPs (B). a1 and a2 represent the bright-field and fluorescence microscopy pictures in the experimental group, respectively. b1 and b2 represent the bright-field and fluorescence microscopy pictures in the control group, respectively. All images were obtained with fluorescence microscopy (100 × oil). Scale bar = 5 μm
The Specificity Verification of SA-FSiNPs
To demonstrate the specificity of SA-FSiNPs for targeted bacteria detection, different concentrations of bacteria mixtures containing bacteria S. typhimurium, E. coli, and B. subtilis were treated with the same amount of nanoprobes. As shown in Fig. 4, a1 and a2 represent the bright-field and fluorescence microscopy pictures of group A bacteria, respectively. b1 and b2 represent the bright-field and fluorescence microscopy pictures of group B bacteria, respectively. c1 and c2 represent the bright-field and fluorescence microscopy pictures of group C bacteria, respectively. A dramatic decrease in the number of bacterial colonies with significant red fluorescence was observed as the bacteria S. typhimurium concentration in the bacteria mixtures decreased. In addition, to further demonstrate the NP probe binds specifically to bacteria S. typhimurium in mixed bacterial samples, pure bacteria S. typhimurium and pure E. coli DH5ɑ and pure B. subtilis were labeled with biotin-labeled bacteria S. typhimurium aptamers and SA-FSiNPs, respectively. Significant red fluorescence in bacterial colonies was observed in the fluorescence image of bacteria S. typhimurium, while no fluorescence was observed in the fluorescence images of E. coli DH5ɑ and B. subtilis (Additional file 1). The results show the great selectivity of nanoprobes for bacteria S. typhimurium labeling, which is due to the specific recognition between the aptamer and bacteria S. typhimurium and no nonspecific binding between the nanoprobes and bacteria.
Photostability of Streptavidin-Conjugated Silica Fluorescence Nanoprobes
It is known that the severe photobleaching of fluorescent dye molecules hinder its application for biological labeling. To verify the anti-photobleaching property of RuBPY dyes doped inside the silica matrix, bacteria S. typhimurium were labeled with SA-FSiNPs and commercially available RBITC-labeled aptamers and excited for 10 min by the successive intense irradiation simultaneously. The fluorescence images were captured at the time gradient of 0 s, 60 s, 2 min, 5 min, and 10 min. As shown in Fig. 5a, the prepared nanoprobes display a significant anti-photobleaching feature. The red signals were clearly distinguishable in the naked eyes even after successive intense irradiation for 10 min. The photostability of the encapsulated fluorescent dye molecules is prominent due to the protective effect of the silica shell, which can prevent from photodamaging oxidation [16, 35, 36]. Quantification of bacterial cell staining was performed using ImageJ software according to its standard manual and the reported method [37, 38]. All the bacterial cells in the images were selected individually using the freehand selection tool of the software. The mean pixel intensity of each bacterial cell on the fluorescence image was measured. Error bars reflect the standard error of the mean. Statistical significance was determined by Student’s t test. As shown in Fig. 5b, the fluorescence of RBITC was bleached quickly during 60-s irradiation.
a Photostability comparison of SA-FSiNPs and RBITC dye. Bacteria S. typhimurium were labeled with the biotin-labeled aptamer and SA-FSiNPs (a1–a5) and RBITC-labeled aptamer (b1–b5), respectively, and excited for 10 min by the successive intense irradiation. The fluorescence images were acquired at 0 s (a1 and b1), 60 s (a2 and b2), 2 min (a3 and b3), 5 min (a4 and b4), and 10 min (a5 and b5), respectively. Scale bar = 5 μm. b Quantitative analysis of bacterial cells, mean ± error bar (set at P < 0.05) versus different probes
Application of the Bacteria S. typhimurium Detection out of Chicken Samples
To verify the feasibility of SA-FSiNPs in the practical application, the chicken samples contaminated with bacteria S. typhimurium were firstly detected using the established aptamer combining the SA-FSiNP treatment. The chicken samples without bacterial contamination process were set as the control groups. As shown in Fig. 6, a1 and a2 represent the bright-field and fluorescence microscopy results in the experimental group, respectively, while the b1 and b2 represent the information of the control group accordingly. A large number of bacterial colonies with significant red fluorescence were clearly distinguished under the fluorescence microscope in the experimental group, which indicates that the bacteria S. typhimurium present in the chicken samples are successfully labeled. In contrast, the control group shows no obvious fluorescence image, implying the control group samples are bacteria S. typhimurium-free. In addition, it is further confirmed that no nonspecific binding occurred between the nanoprobes and other potential bacteria in the chicken samples. So, these aptamer-based SA-FSiNPs are probably operative in the normal surveillance task, which is yet to be verified.
Fluorescence images of bacteria S. typhimurium treated with the biotin-labeled aptamer and SA-FSiNPs in the polluted chicken samples (A) and in the normal chicken samples (B). a1 and a2 represent the bright-field and fluorescence microscopy pictures in the experimental group, respectively. b1 and b2 represent the bright-field and fluorescence microscopy pictures in the control group, respectively. All images were obtained with fluorescence microscopy (100 × oil). Scale bar = 5 μm
Conclusions
A simple but effective method based on biotin-labeled aptamer and streptavidin-conjugated silica fluorescence nanoprobes for bacteria S. typhimurium detection had been established. The biotin-labeled anti-bacteria S. typhimurium aptamer shows high selectivity and affinity to membrane proteins of this food-borne pathogen. Because of the specific binding between streptavidin and biotin, the SA-FSiNPs can be effectively applied for the labeling of bacteria S. typhimurium in various complex backgrounds, which had been further confirmed in the multiple bacteria mixture and practical chicken samples. In addition, SA-FSiNPs display good photostability than the dye-labeled probes. Moreover, silica nanomaterials take advantage of good biocompatibility, which can serve as a necessary and useful supplement for the current detection methods.
References
Suo ZY, Yang XH, Avci R, Kellerman L, Pascual DW, Fries M et al (2007) HEPES-stabilized encapsulation of Salmonella typhimurium. Langmuir 23(3):1365–1374
Li J, Nakayasu ES, Overall CC, Johnson RC, Kidwai AS, McDermott JE et al (2015) Global analysis of Salmonella alternative sigma factor E on protein translation. J Proteome Res 14(4):1716–1726
Mitchell E, O’Mahony M, Lynch D, Ward LR, Rowe B, Uttley A et al (1989) Large outbreak of food poisoning caused by Salmonella typhimurium definitive type 49 in mayonnaise. Br Med J 298(6666):99–101
Domingues S, Matos RG, Reis FP, Fialho AM, Barbas A, Arraiano CM (2009) Biochemical characterization of the RNase II family of exoribonucleases from the human pathogens Salmonella typhimurium and Streptococcus pneumoniae. Biochem 48(50):11848–11857
Magliulo M, Simoni P, Guardigli M, Michelini E, Luciani M, Lelli R et al (2007) A rapid multiplexed chemiluminescent immunoassay for the detection of Escherichia coli O157:H7, Yersinia enterocolitica, Salmonella typhimurium, and Listeria monocytogenes pathogen bacteria. J Agric Food Chem 55(13):4933–4939
Jyoti A, Vajpayee P, Singh G, Patel CB, Gupta KC, Shanker R (2011) Identification of environmental reservoirs of nontyphoidal salmonellosis: aptamer-assisted bioconcentration and subsequent detection of Salmonella typhimurium by quantitative polymerase chain reaction. Environ Sci Technol 45(20):8996–9002
Medley CD, Bamrungsap S, Tan WH, Smith JE (2011) Aptamer-conjugated nanoparticles for cancer cell detection. Anal Chem 83(3):727–734
Labib M, Zamay AS, Kolovskaya OS, Reshetneva IT, Zamay GS, Kibbee RJ et al (2012) Aptamer-based viability impedimetric sensor for bacteria. Anal Chem 84(21):8966–8969
Rhinehardt KL, Srinivas G, Mohan RV (2015) Molecular dynamics simulation analysis of anti-MUC1 aptamer and mucin 1 peptide binding. J Phys Chem B 119(22):6571–6583
Armstrong RE, Strouse GF (2014) Rationally manipulating aptamer binding affinities in a stem-loop molecular beacon. Bioconjugate Chem 25(10):1769–1776
Kim B, Jung IH, Kang M, Shim HK, Woo HY (2012) Cationic conjugated polyelectrolytes-triggered conformational change of molecular beacon aptamer for highly sensitive and selective potassium ion detection. J Am Chem Soc 134(6):3133–3138
Joshi R, Janagama H, Dwivedi HP, Senthil Kumar TMA, Jaykus LA, Schefers J et al (2009) Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol Cell Probes 23(1):20–28
Book B, Chen JJ, Irudayaraj J (2011) Quantification of receptor targeting aptamer binding characteristics using single-molecule spectroscopy. Biotechnol Bioeng 108(5):1222–1227
Duan N, Wu SJ, Chen XJ, Huang YK, Xia Y, Ma XY et al (2013) Selection and characterization of aptamers against Salmonella typhimurium using whole-bacterium systemic evolution of ligands by exponential enrichment (SELEX). J Agric Food Chem 61(13):3229–3234
Yao J, Yang M, Duan YX (2014) Chemistry, biology, and medicine of fluorescent nanomaterials and related systems: new insights into biosensing, bioimaging, genomics, diagnostics, and therapy. Chem Rev 114(12):6130–6178
Lian W, Litherland SA, Badrane H, Tan WH, Wu DH, Baker HV (2004) Ultrasensitive detection of biomolecules with fluorescent dye-doped nanoparticles. Anal Biochem 334(1):135–144
Lee JE, Lee N, Kim H, Kim J, Choi SH, Kim JH et al (2010) Uniform mesoporous dye-doped silica nanoparticles decorated with multiple magnetite nanocrystals for simultaneous enhanced magnetic resonance imaging, fluorescence imaging, and drug delivery. J Am Chem Soc 132(2):552–557
Wang KM, He XX, Yang XH, Shi H (2013) Functionalized silica nanoparticles: a platform for fluorescence imaging at the cell and small animal levels. Acc Chem Res 46(7):1367–1376
Shi H, He XX, Yuan Y, Wang KM, Tan WH (2010) Nanoparticle-based biocompatible and long-Life marker for lysosome labeling and tracking. Anal Chem 82(6):2213–2220
Song X, Li F, Ma JW, Jia NQ, Xu JM, Shen HB (2011) Synthesis of fluorescent silica nanoparticles and their applications as fluorescence probes. J Fluoresc 21(3):1205–1212
Chen ZZ, Cai L, Chen MY, Lin Y, Pang DW, Tang HW (2015) Indirect immunofluorescence of E. coli O157:H7 with fluorescent silica nanoparticles. Biosens Bioelectron 66:95–102
Liu AH, Wu LY, He ZL, Zhou JZ (2011) Development of highly fluorescent silica nanoparticles chemically doped with organic dye for sensitive DNA microarray detection. Anal Bioanal Chem 401(6):2003–2011
Rossi LM, Shi LF, Quina FH, Rosenzweig Z (2005) Stöber synthesis of monodispersed luminescent silica nanoparticles for bioanalytical assays. Langmuir 21(10):4277–4280
Hun X, Zhang Z (2007) Functionalized fluorescent core-shell nanoparticles used as a fluorescent labels in fluoroimmunoassay for IL-6. Biosens Bioelectron 22(11):2743–2748
Du Y, Li BL, Wang EK (2013) “Fitting” makes “sensing” simple: label-free detection strategies based on nucleic acid aptamers. Acc Chem Res 46(2):203–213
Yi M, Yang S, Peng ZY, Liu CH, Li JS, Zhong WW et al (2014) Two-photon graphene oxide/aptamer nanosensing conjugate for in vitro or in vivo molecular probing. Anal Chem 86(7):3548–3554
Tan LH, Xing H, Lu Y (2014) DNA as a powerful tool for morphology control, spatial positioning, and dynamic assembly of nanoparticles. Acc Chem Res 47(6):1881–1890
Tan WH, Donovan MJ, Jiang JH (2013) Aptamers from cell-based selection for bioanalytical applications. Chem Rev 113(4):2842–2862
Arriagada FJ, Osseo-Asare K (1999) Synthesis of nanosize silica in a nonionic water-in-oil microemulsion: effects of the water/surfactant molar ratio and ammonia concentration. J Colloid Interface Sci 211(2):210–220
Cai L, Chen ZZ, Chen MY, Tang HW, Pang DW (2013) MUC-1 aptamer-conjugated dye-doped silica nanoparticles for MCF-7 cells detection. Biomaterials 34(2):371–381
Lee JE, Lee NY, Kim T, Kim JY, Hyeon TW (2011) Multifunctional mesoporous silica nanocomposite nanoparticles for theranostic applications. Acc Chem Res 44(10):893–902
Kim ST, Kim DJ, Kim TJ, Seo DW, Kim TH, Lee SY et al (2010) Novel streptavidin-functionalized silicon nanowire arrays for CD4+ T lymphocyte separation. Nano Lett 10(8):2877–2883
Schwizer F, Köhler V, Dürrenberger M, Knörr L, Ward TR (2013) Genetic optimization of the catalytic efficiency of artificial imine reductases based on biotin-streptavidin technology. ACS Catal 3(8):1752–1755
Shukla RS, Tai W, Mahato R, Jin W, Cheng K (2013) Development of streptavidin-based nanocomplex for siRNA delivery. Mol Pharmaceutics 10(12):4534–4545
Lal M, Levy L, Kim KS, He GS, Wang X, Min YH et al (2000) Silica nanobubbles containing an organic dye in a multilayered organic/inorganic heterostructure with enhanced luminescence. Chem Mater 12(9):2632–2639
Zhao XJ, Bagwe RP, Tan WH (2004) Development of organic-dye-doped silica nanoparticles in a reverse microemulsion. Adv Mater 16(2):173–176
Liu F, Ni ASY, Lim Y, Mohanram H, Bhattacharjya S, Xing BG (2012) Lipopolysaccharide neutralizing peptide-porphyrin conjugates for effective photoinactivation and intracellular imaging of gram-negative bacteria strains. Bioconjugate Chem 23(8):1639–1647
Hamliton N (2009) Quantification and its application in fluorescent microscopy imaging. Traffic 10(8):951–961
Acknowledgements
This work was financially supported by the Fundamental Research Funds for the Central Universities (No. JUSRP11568), a grant from Public Health Research Center at Jiangnan University (No. JUPH201505), and a grant from Jiangsu Science and Technology Department (No. BM2015024-3).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing Interests
The authors declare that they have no competing interests.
Authors’ Contributions
QYW and YJK designed the experiments. QYW performed the experiments. QYW and YJK analyzed the data and wrote the manuscript. Both authors read and approved the final manuscript.
Authors’ Information
QYW is a teaching assistant and experimentalist in the College of Laboratory Medicine, Hunan University of Medicine, Huaihua, Hunan, People’s Republic of China. YJK is a lecturer in Wuxi Medical School and Public Health Research Center, Jiangnan University, Wuxi, Jiangsu, People’s Republic of China.
Additional file
Additional file 1:
The results of bacterial detection tests in terms of higher concentrated samples. (DOC 864 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, QY., Kang, YJ. Bioprobes Based on Aptamer and Silica Fluorescent Nanoparticles for Bacteria Salmonella typhimurium Detection. Nanoscale Res Lett 11, 150 (2016). https://doi.org/10.1186/s11671-016-1359-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s11671-016-1359-z